Introducing GRAViTy-V2: Advancing viral taxonomy
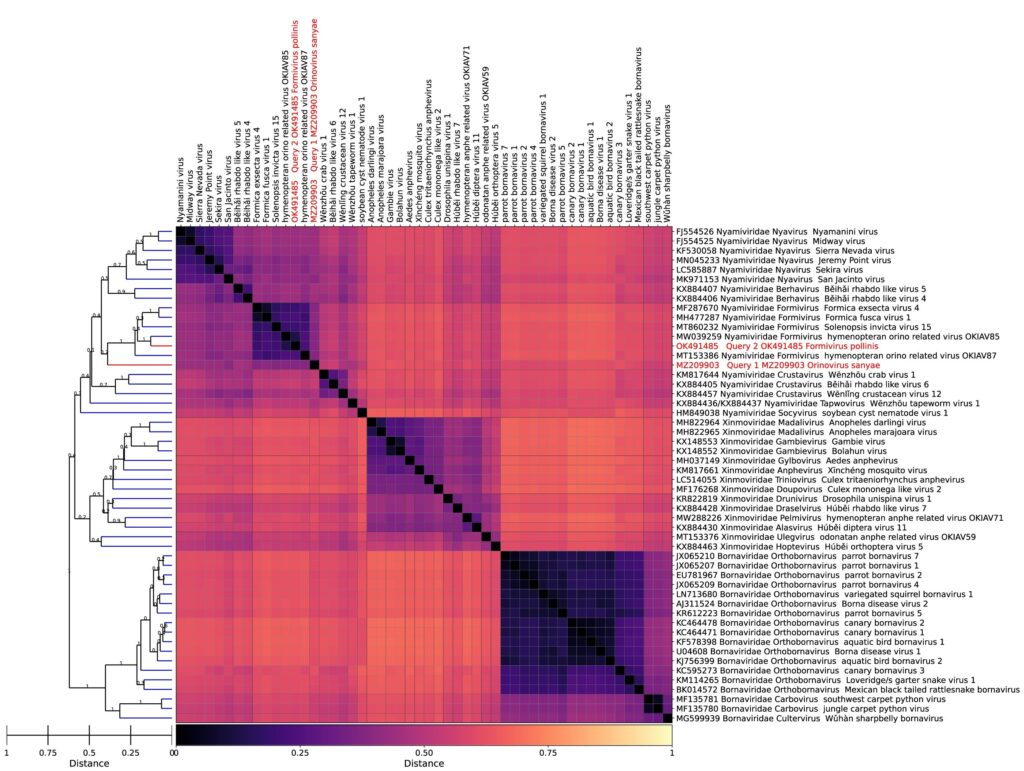
In collaboration with the University of Oxford, the University of the West of England, and Quadram Institute Bioscience, we are excited to introduce GRAViTy-V2, an advanced bioinformatics tool for viral taxonomy research.
Building upon our “Genome Relationship Applied to Virus Taxonomy” (GRAViTy) framework, GRAViTy-V2 is specifically designed to classify viruses using coding-complete genome sequence data.
Challenges of virus classification
Viral taxonomy is now becoming increasingly relying on evolutionary relatedness; however, viruses, as some of the fastest-evolving biological entities on this planet, often display significant dissimilarity across much of their genomes. To estimate evolutionary relationships from molecular data, most standard tools require sequences with detectable similarities across all viruses under analysis. Consequently, researchers are often limited to analysing only some of the most conserved genomic regions, discarding other that, while not universally conserved, could still be highly informative and provide valuable insights for that specific subsets.
This limitation is particularly problematic when analysing diverse groups of viruses, such as those spanning an entire viral family or order.
The GRAViTy framework: A novel solution
Our GRAViTy framework was developed to address precisely these challenges. It transforms each genome into a protein-coding region profile, and calculates a single metric, termed the Composite Generalised Jaccard Similarity index, or Composite Jaccard Score in short, to measure overall similarity between these profiles, which can then be used to approximate evolutionary relationships and in turn classify viruses. The score incorporates not only the presence or absence of protein-coding regions, but also their sequence similarity, locations, orders, and orientations.
This method allows researchers to utilise the full breadth of genomic information for virus classification, and at the same time bypasses the need for detectable similarity across all viruses, enabling broader and more inclusive virus taxonomy research.
Advancements in GRAViTy-V2
The initial implementation of GRAViTy, while innovative, was computationally demanding and lacked user-friendly packaging. GRAViTy-V2 addresses these issues with significant updates and optimisations:
Streamlined and accessible: GRAViTy-V2 is now a standalone, user-friendly application.
Enhanced outputs: GRAViTy-V2 now gives a range of ‘explainability’ features for describing how classifications are made, supporting a wider range of taxonomy tasks.
Improved performance: Optimisations enable efficient analysis of large datasets, including those with many previously unclassified sequences.
GRAViTy-V2 has demonstrated results comparable to expert-curated classifications, validated against datasets from the International Committee on Taxonomy of Viruses.
Unlocking new possibilities
GRAViTy-V2 offers a robust tool for exploring viral diversity, aiding researchers in understanding virus evolution and streamlining taxonomy workflows. The software is available as an open-source package, with detailed installation instructions and workflows accessible through this GitHub repository.
We invite you to explore its potential and use this powerful tool in your research!
Pakorn Aiewsakun
Co-author

